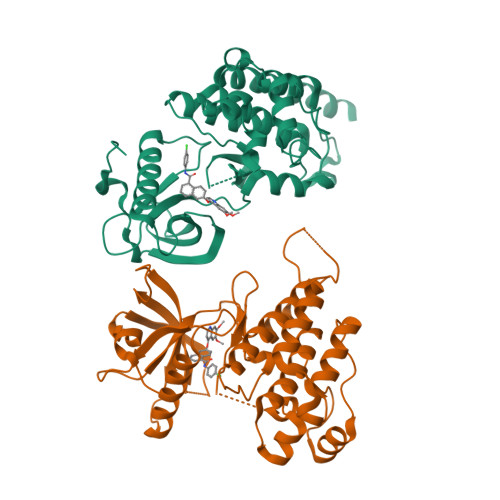
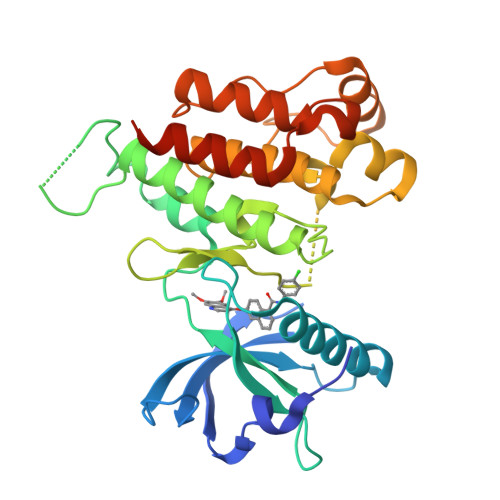
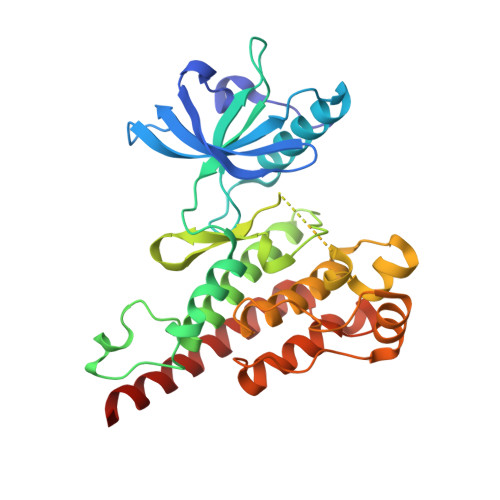
Naphthamides as Novel and Potent Vascular Endothelial Growth Factor Receptor Tyrosine Kinase Inhibitors: Design, Synthesis and Evaluation
Harmange, J.C., Weiss, M.M., Germain, J., Polverino, A.J., Borg, G., Bready, J., Chen, D., Choquette, D., Coxon, A., DeMelfi, T., DiPietro, L., Doerr, N., Estrada, J., Flynn, J., Graceffa, R.F., Harriman, S.P., Kaufman, S., La, D.S., Long, A., Martin, M.W., Neervannan, S., Patel, V.F., Potashman, M., Regal, K., Roveto, P.M., Schrag, M.L., Starnes, C., Tasker, A., Teffera, Y., Wang, L., White, R.D., Whittington, D.A., Zanon, R.(2008) J Med Chem 51: 1649-1667
- PubMed: 18324761
- DOI: https://doi.org/10.1021/jm701097z
- Primary Citation of Related Structures:
3B8Q, 3BE2 - PubMed Abstract:
A series of naphthyl-based compounds were synthesized as potential inhibitors of vascular endothelial growth factor (VEGF) receptors. Investigations of structure-activity relationships led to the identification of a series of naphthamides that are potent inhibitors of the VEGF receptor tyrosine kinase family. Numerous analogues demonstrated low nanomolar inhibition of VEGF-dependent human umbilical vein endothelial cell (HUVEC) proliferation, and of these several compounds possessed favorable pharmacokinetic (PK) profiles. In particular, compound 48 demonstrated significant antitumor efficacy against established HT29 human colon adenocarcinoma xenografts implanted in athymic mice. A full account of the preparation, structure-activity relationships, pharmacokinetic properties, and pharmacology of analogues within this series is presented.
Organizational Affiliation:
Department of Medicinal Chemistry, Amgen Inc., One Kendall Square, Cambridge, MA 02139, USA. harmange@amgen.com